3D printable CNVpytor logo (stl file)
CNVpytor is a Python package and command line tool for CNV/CNA analysis from depth-of-coverage by mapped reads developed in Abyzov Lab, Mayo Clinic.
Follow CNVpytor Twitter account.
CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing
Milovan Suvakov, Arijit Panda, Colin Diesh, Ian Holmes, Alexej Abyzov, GigaScience, Volume 10, Issue 11, November 2021, giab074
https://doi.org/10.1093/gigascience/giab074
- Geting started with command line interface
- Jupyter notebook: How to use CNVpytor from Python
- Google Colab: With CEPH trio example dataset
- Video Tutorial: 3-minute YT demo
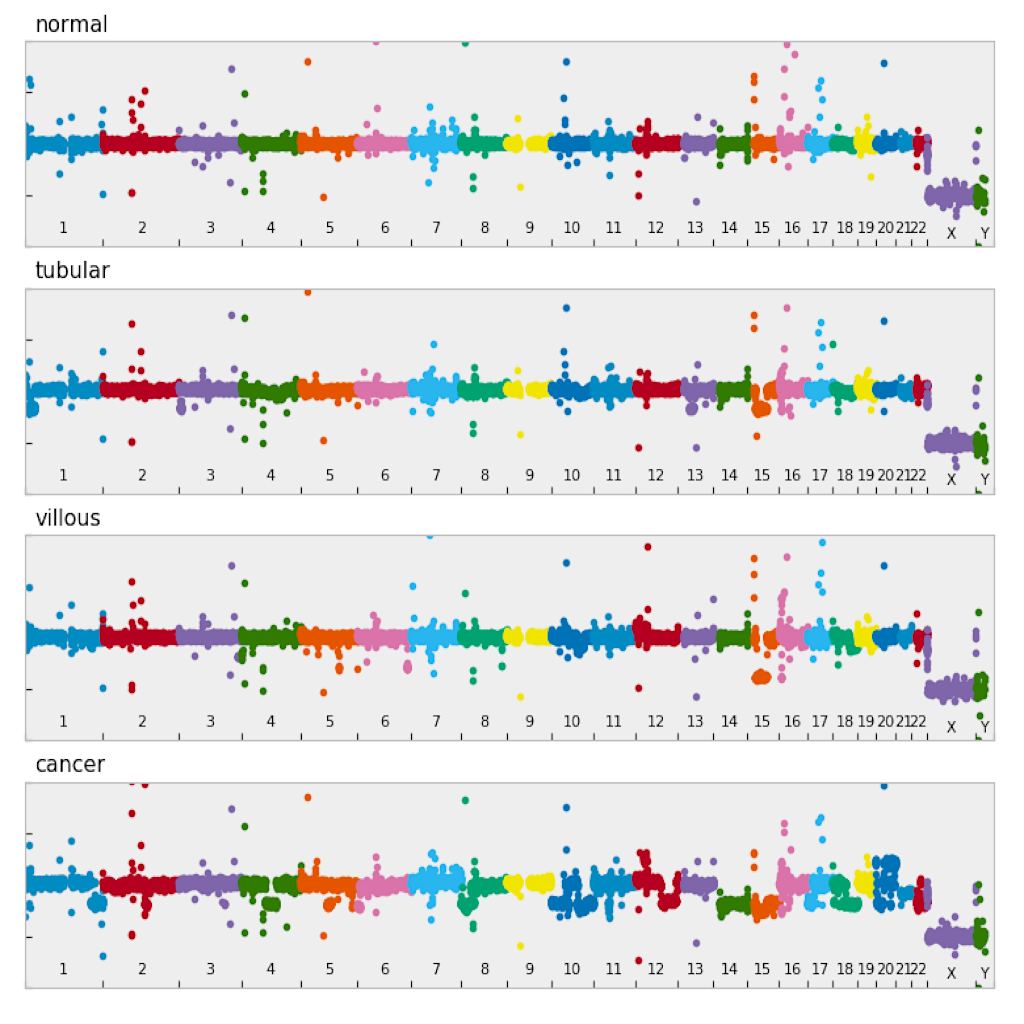
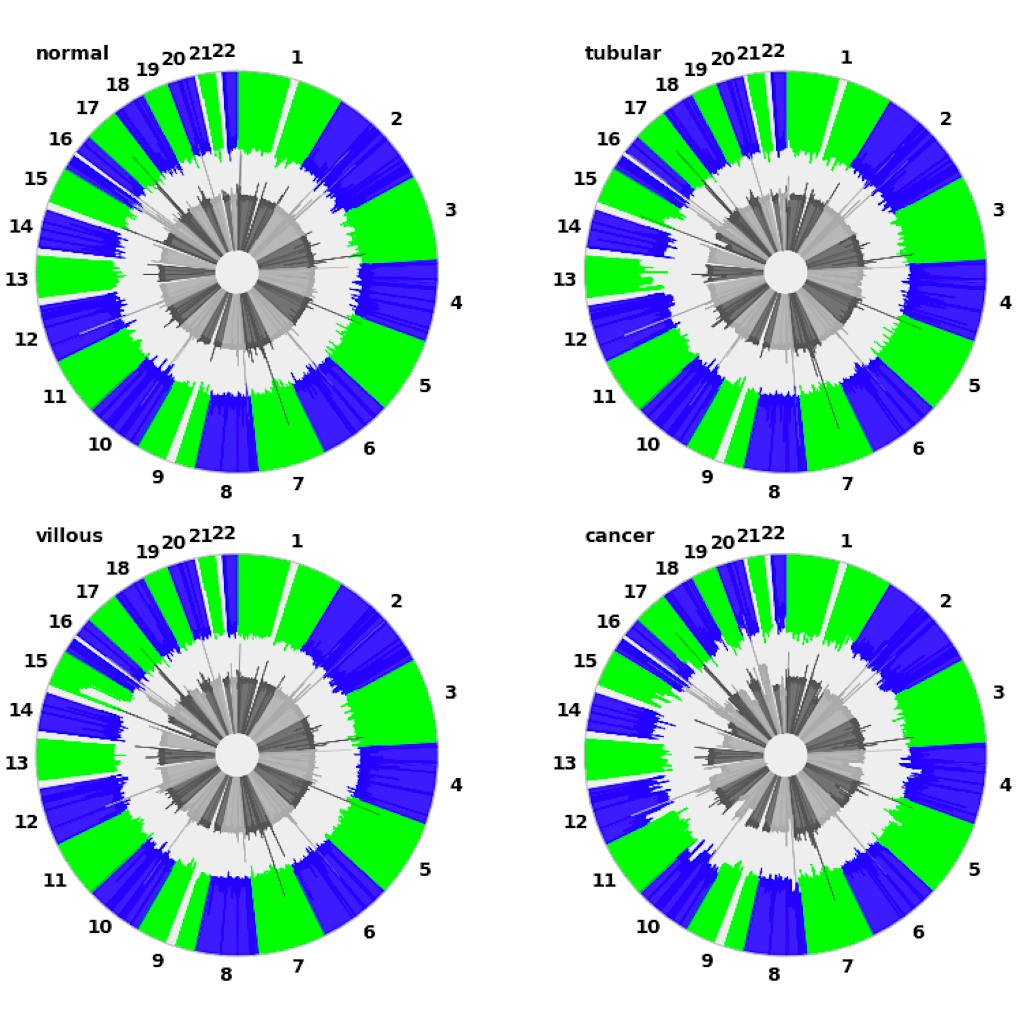
| Manhattan plot (see example) | Circular plot (see example) |
 |
 |
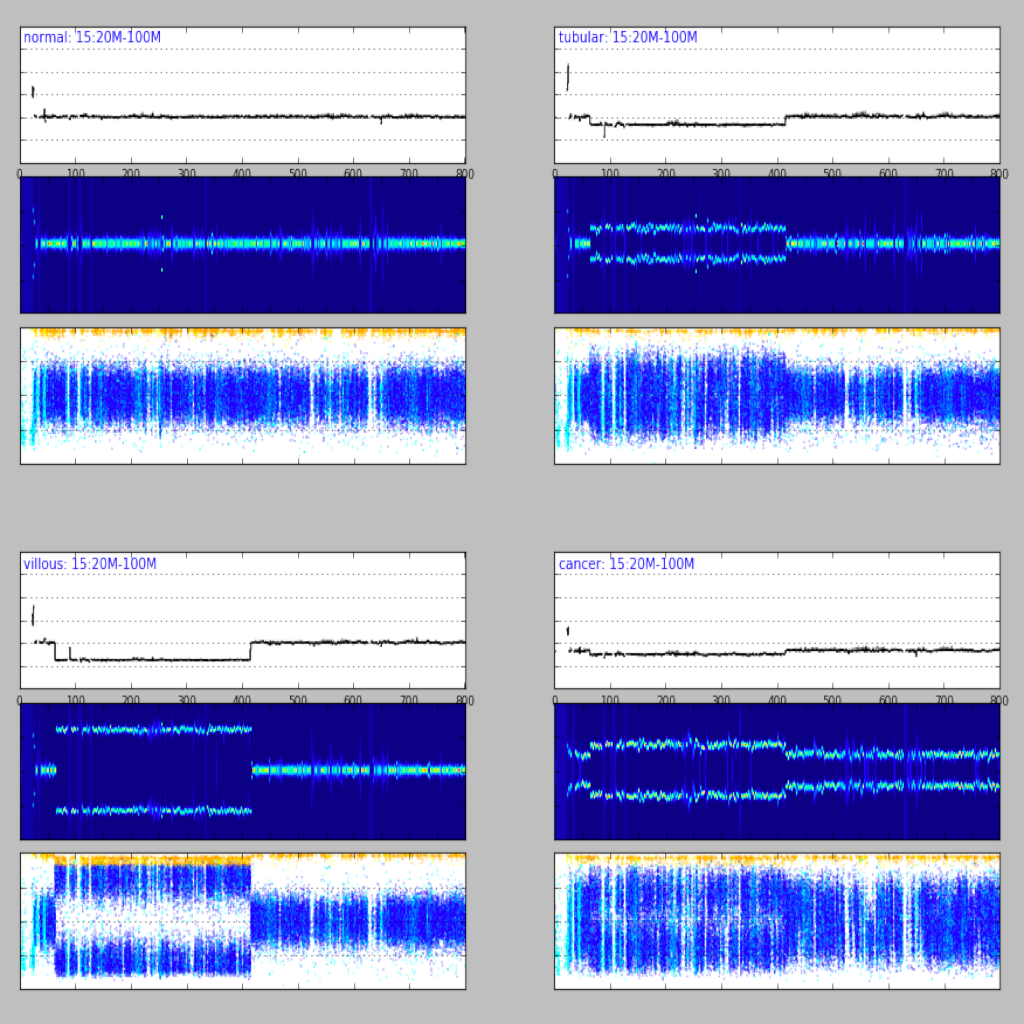
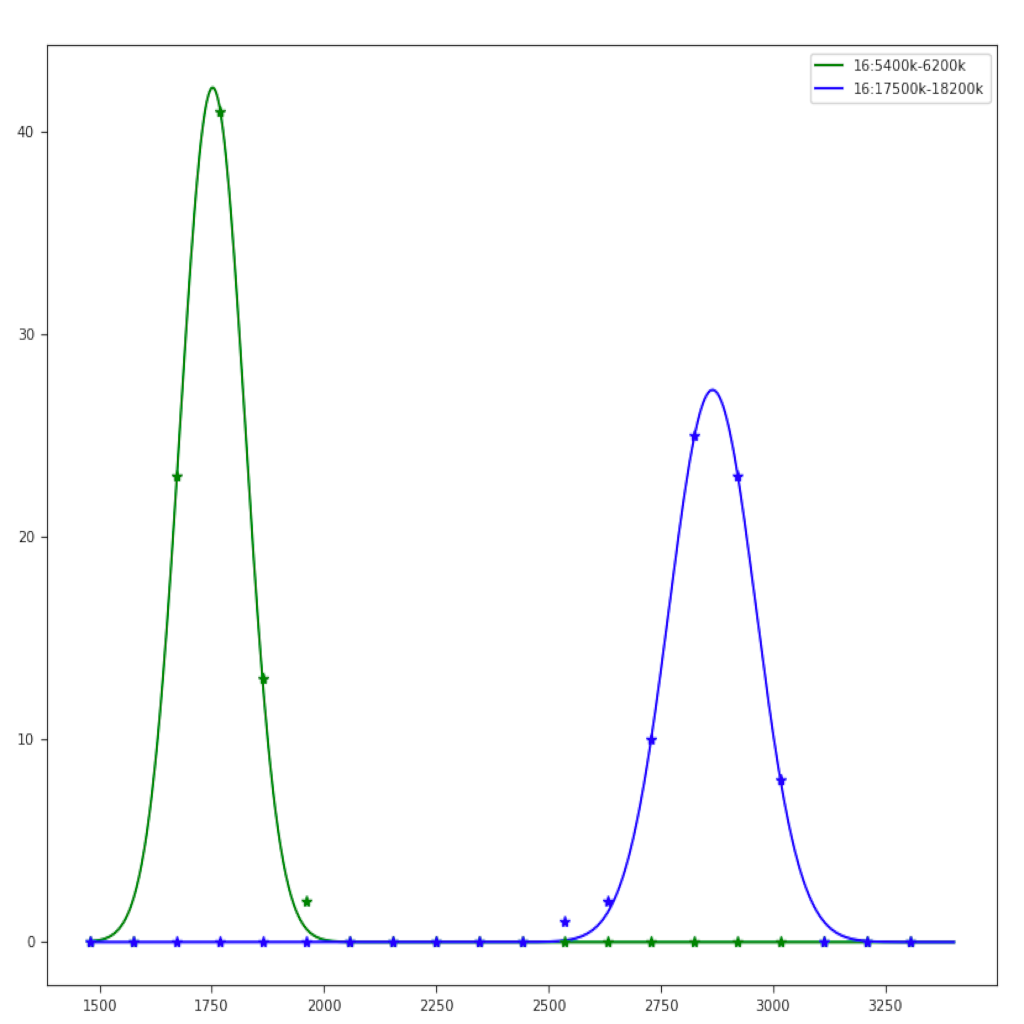
| Region plot (see example) | Compare regions (see example) |
 |
 |
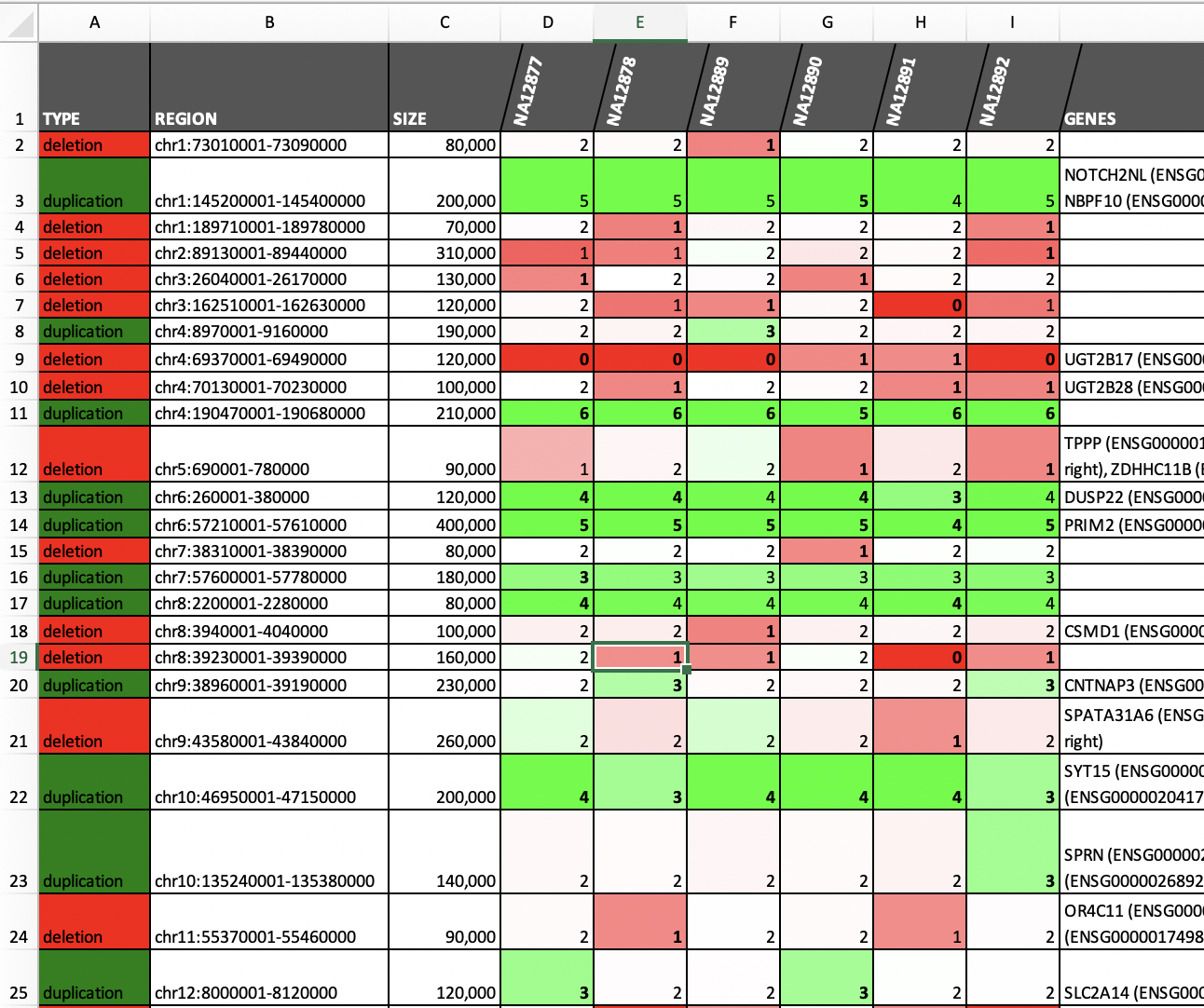
| Merging and annotating calls (see example) | Call somatic CNAs (see example) |
 |
 |
> pip install cnvpytor
> cnvpytor -download
Check CNVpytor github page.
Released under MIT licence.