-
Notifications
You must be signed in to change notification settings - Fork 6
/
Copy path07b_pythia_xarray_comp.py
412 lines (233 loc) · 12.4 KB
/
07b_pythia_xarray_comp.py
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
#!/usr/bin/env python
# coding: utf-8
# # Computations and Masks with Xarray
# ---
# ## Imports
#
# In[ ]:
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
# Let's open the monthly sea surface temperature (SST) data from the Community Earth System Model v2 (CESM2), which is a Global Climate Model:
# In[ ]:
ds = xr.open_dataset('data/CESM2_sst_data.nc')
ds
# ## Arithmetic Operations
#
# Arithmetic operations with a single DataArray automatically apply over all array values (like NumPy). This process is called vectorization. Let's convert the air temperature from degrees Celsius to kelvins:
# In[ ]:
ds.tos + 273.15
# Lets's square all values in `tos`:
# In[ ]:
ds.tos**2
# ## Aggregation Methods
#
# A very common step during data analysis is to summarize the data in question by computing aggregations like `sum()`, `mean()`, `median()`, `min()`, `max()` in which reduced data provide insight into the nature of large dataset. Let's explore some of these aggregation methods.
#
# Compute the mean:
# In[ ]:
ds.tos.mean()
# Because we specified no `dim` argument the function was applied over all dimensions, computing the mean of every element of `tos` across time and space. It is possible to specify a dimension along which to compute an aggregation. For example, to calculate the mean in time for all locations, specify the time dimension as the dimension along which the mean should be calculated:
# In[ ]:
ds.tos.mean(dim='time').plot(size=7);
# Compute the temporal min:
# In[ ]:
ds.tos.min(dim=['time'])
# Compute the spatial sum:
# In[ ]:
ds.tos.sum(dim=['lat', 'lon'])
# Compute the temporal median:
# In[ ]:
ds.tos.median(dim='time')
# The following table summarizes some other built-in xarray aggregations:
#
# | Aggregation | Description |
# |--------------------------|---------------------------------|
# | ``count()`` | Total number of items |
# | ``mean()``, ``median()`` | Mean and median |
# | ``min()``, ``max()`` | Minimum and maximum |
# | ``std()``, ``var()`` | Standard deviation and variance |
# | ``prod()`` | Compute product of elements |
# | ``sum()`` | Compute sum of elements |
# | ``argmin()``, ``argmax()``| Find index of minimum and maximum value |
# ## GroupBy: Split, Apply, Combine
#
# Simple aggregations can give useful summary of our dataset, but often we would prefer to aggregate conditionally on some coordinate labels or groups. Xarray provides the so-called `groupby` operation which enables the **split-apply-combine** workflow on xarray DataArrays and Datasets. The split-apply-combine operation is illustrated in this figure
#
# <img src="./images/xarray-split-apply-combine.jpeg">
#
# This makes clear what the `groupby` accomplishes:
#
# - The split step involves breaking up and grouping an xarray Dataset or DataArray depending on the value of the specified group key.
# - The apply step involves computing some function, usually an aggregate, transformation, or filtering, within the individual groups.
# - The combine step merges the results of these operations into an output xarray Dataset or DataArray.
#
# We are going to use `groupby` to remove the seasonal cycle ("climatology") from our dataset. See the [xarray `groupby` user guide](https://xarray.pydata.org/en/stable/user-guide/groupby.html) for more examples of what `groupby` can take as an input.
# First, let's select a gridpoint closest to a specified lat-lon, and plot a time series of SST at that point. The annual cycle will be quite evident.
# In[ ]:
ds.tos.sel(lon=310, lat=50, method='nearest').plot();
# ### Split
#
# Let's group data by month, i.e. all Januaries in one group, all Februaries in one group, etc.
#
# In[ ]:
ds.tos.groupby(ds.time.dt.month)
# <div class="admonition alert alert-info">
#
# In the above code, we are using the `.dt` [`DatetimeAccessor`](https://xarray.pydata.org/en/stable/generated/xarray.core.accessor_dt.DatetimeAccessor.html) to extract specific components of dates/times in our time coordinate dimension. For example, we can extract the year with `ds.time.dt.year`. See also the equivalent [Pandas documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.dt.html).
#
# </div>
# Xarray also offers a more concise syntax when the variable you’re grouping on is already present in the dataset. This is identical to `ds.tos.groupby(ds.time.dt.month)`:
# In[ ]:
ds.tos.groupby('time.month')
# ### Apply & Combine
#
# Now that we have groups defined, it’s time to “apply” a calculation to the group. These calculations can either be:
#
# - aggregation: reduces the size of the group
# - transformation: preserves the group’s full size
#
# At then end of the apply step, xarray will automatically combine the aggregated/transformed groups back into a single object.
#
#
#
# #### Compute climatology
#
#
# Let's calculate the climatology at every point in the dataset:
#
# In[ ]:
tos_clim = ds.tos.groupby('time.month').mean()
tos_clim
# Plot climatology at a specific point:
# In[ ]:
tos_clim.sel(lon=310, lat=50, method='nearest').plot();
# Plot zonal mean climatology:
# In[ ]:
tos_clim.mean(dim='lon').transpose().plot.contourf(levels=12, cmap='turbo');
# Calculate and plot the difference between January and December climatologies:
# In[ ]:
(tos_clim.sel(month=1) - tos_clim.sel(month=12)).plot(size=6, robust=True);
# #### Compute anomaly
#
# Now let's combine the previous steps to compute climatology and use xarray's `groupby` arithmetic to remove this climatology from our original data:
# In[ ]:
gb = ds.tos.groupby('time.month')
tos_anom = gb - gb.mean(dim='time')
tos_anom
# In[ ]:
tos_anom.sel(lon=310, lat=50, method='nearest').plot();
# Let's compute and visualize the mean global anomaly over time. We need to specify both `lat` and `lon` dimensions in the `dim` argument to `mean()`:
# In[ ]:
unweighted_mean_global_anom = tos_anom.mean(dim=['lat', 'lon'])
unweighted_mean_global_anom.plot();
# <div class="admonition alert alert-warning">
#
#
# An operation which combines grid cells of different size is not scientifically valid unless each cell is weighted by the size of the grid cell. Xarray has a convenient [`.weighted()`](https://xarray.pydata.org/en/stable/user-guide/computation.html#weighted-array-reductions) method to accomplish this
#
# </div>
#
# Let's first load the cell area data from another CESM2 dataset that contains the weights for the grid cells:
# In[ ]:
filepath2 = DATASETS.fetch('CESM2_grid_variables.nc')
areacello = xr.open_dataset(filepath2).areacello
areacello
# As before, let's calculate area-weighted mean global anomaly:
# In[ ]:
weighted_mean_global_anom = tos_anom.weighted(areacello).mean(dim=['lat', 'lon'])
# Let's plot both unweighted and weighted means:
# In[ ]:
unweighted_mean_global_anom.plot(size=7)
weighted_mean_global_anom.plot()
plt.legend(['unweighted', 'weighted']);
# ## Other high level computation functionality
#
# - `resample`: [Groupby-like functionality specifialized for time dimensions. Can be used for temporal upsampling and downsampling](https://xarray.pydata.org/en/stable/user-guide/time-series.html#resampling-and-grouped-operations)
# - `rolling`: [Useful for computing aggregations on moving windows of your dataset e.g. computing moving averages](https://xarray.pydata.org/en/stable/user-guide/computation.html#rolling-window-operations)
# - `coarsen`: [Generic functionality for downsampling data](https://xarray.pydata.org/en/stable/user-guide/computation.html#coarsen-large-arrays)
#
#
# For example, resample to annual frequency:
# In[ ]:
r = ds.tos.resample(time='AS')
r
# In[ ]:
r.mean()
# Compute a 5-month moving average:
# In[ ]:
m_avg = ds.tos.rolling(time=5, center=True).mean()
m_avg
# In[ ]:
lat = 50
lon = 310
m_avg.isel(lat=lat, lon=lon).plot(size=6)
ds.tos.isel(lat=lat, lon=lon).plot()
plt.legend(['5-month moving average', 'monthly data']);
# ## Masking Data
#
# Using the `xr.where()` or `.where()` method, elements of an xarray Dataset or xarray DataArray that satisfy a given condition or multiple conditions can be replaced/masked. To demonstrate this, we are going to use the `.where()` method on the `tos` DataArray.
# We will use the same sea surface temperature dataset:
# In[ ]:
ds
# ### Using `where` with one condition
# Imagine we wish to analyze just the last time in the dataset. We could of course use `.isel()` for this:
# In[ ]:
sample = ds.tos.isel(time=-1)
sample
# Unlike `.isel()` and `.sel()` that change the shape of the returned results, `.where()` preserves the shape of the original data. It accomplishes this by returning values from the original DataArray or Dataset if the `condition` is `True`, and fills in values (by default `nan`) wherever the `condition` is `False`.
#
# Before applying it, let's look at the [`.where()` documentation](http://xarray.pydata.org/en/stable/generated/xarray.DataArray.where.html). As the documention points out, the conditional expression in `.where()` can be:
#
# - a DataArray
# - a Dataset
# - a function
#
# For demonstration purposes, let's use `.where()` to mask locations with temperature values greater than `0`:
# In[ ]:
masked_sample = sample.where(sample < 0.0)
masked_sample
# Let's plot both our original sample, and the masked sample:
# In[ ]:
fig, axes = plt.subplots(ncols=2, figsize=(19, 6))
sample.plot(ax=axes[0])
masked_sample.plot(ax=axes[1]);
# ### Using `where` with multiple conditions
# `.where()` allows providing multiple conditions. To do this, we need to make sure each conditional expression is enclosed in `()`. To combine conditions, we use the `bit-wise and` (`&`) operator and/or the `bit-wise or` (`|`). Let's use `.where()` to mask locations with temperature values less than 25 and greater than 30:
# In[ ]:
sample.where((sample > 25) & (sample < 30)).plot(size=6);
# We can use coordinates to apply a mask as well. Below, we use the `latitude` and `longitude` coordinates to mask everywhere outside of the [Niño 3.4 region](https://www.ncdc.noaa.gov/teleconnections/enso/indicators/sst/):
#
# 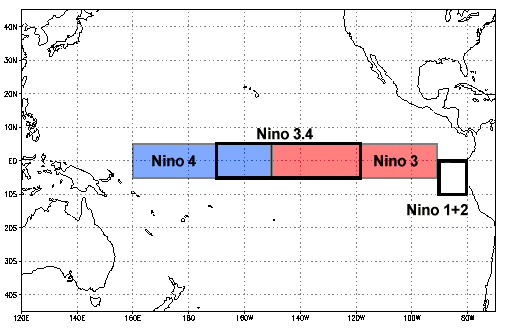
#
#
# In[ ]:
sample.where(
(sample.lat < 5) & (sample.lat > -5) & (sample.lon > 190) & (sample.lon < 240)
).plot(size=6);
# ### Using `where` with a custom fill value
# `.where()` can take a second argument, which, if supplied, defines a fill value for the masked region. Below we fill masked regions with a constant `0`:
# In[ ]:
sample.where((sample > 25) & (sample < 30), 0).plot(size=6);
# ---
# ## Summary
#
# - Similar to NumPy, arithmetic operations are vectorized over a DataArray
# - Xarray provides aggregation methods like `sum()` and `mean()`, with the option to specify which dimension over which the operation will be done
# - `groupby` enables the convenient split-apply-combine workflow
# - The `.where()` method allows for filtering or replacing of data based on one or more provided conditions
#
# ### What's next?
#
# In the next notebook, we will work through an example of plotting the [Niño 3.4 Index](https://climatedataguide.ucar.edu/climate-data/nino-sst-indices-nino-12-3-34-4-oni-and-tni).
# ## Resources and References
#
# - `groupby`: [Useful for binning/grouping data and applying reductions and/or transformations on those groups](https://xarray.pydata.org/en/stable/user-guide/groupby.html)
# - `resample`: [Groupby-like functionality specifialized for time dimensions. Can be used for temporal upsampling and downsampling](https://xarray.pydata.org/en/stable/user-guide/time-series.html#resampling-and-grouped-operations)
# - `rolling`: [Useful for computing aggregations on moving windows of your dataset e.g. computing moving averages](https://xarray.pydata.org/en/stable/user-guide/computation.html#rolling-window-operations)
# - `coarsen`: [Generic functionality for downsampling data](https://xarray.pydata.org/en/stable/user-guide/computation.html#coarsen-large-arrays)
#
# - `weighted`: [Useful for weighting data before applying reductions](https://xarray.pydata.org/en/stable/user-guide/computation.html#weighted-array-reductions)
#
# - [More xarray tutorials and videos](https://xarray.pydata.org/en/stable/tutorials-and-videos.html)
# - [Xarray Documentation - Masking with `where()`](https://xarray.pydata.org/en/stable/user-guide/indexing.html#masking-with-where)